GSoC:MSA
Improvements including Multiple Sequence Alignment Algorithms
Google Summer of Code Project by Mark Chapman
Mentored by Andreas Prlic
Co-mentored by Scooter Willis and Kyle Ellrott
Biologists infer evolutionary, structural, and functional relationships between biopolymers from similarities and divergences of primary structure in multiple sequence alignments. This summer project will add a module which manages an alignment and offers several implementation options. Initial code will establish a framework for alignments that outlines a standard four stage approach of pairwise similarity calculation, hierarchical clustering, progressive alignment, and refinement. Each step will get filled in with details from the most pervasive progressive alignment algorithm, clustalw. Further work will then add alternative methods which update the stages to increase speed, improve accuracy, or change the scope of the multiple sequence alignment.
Milestones
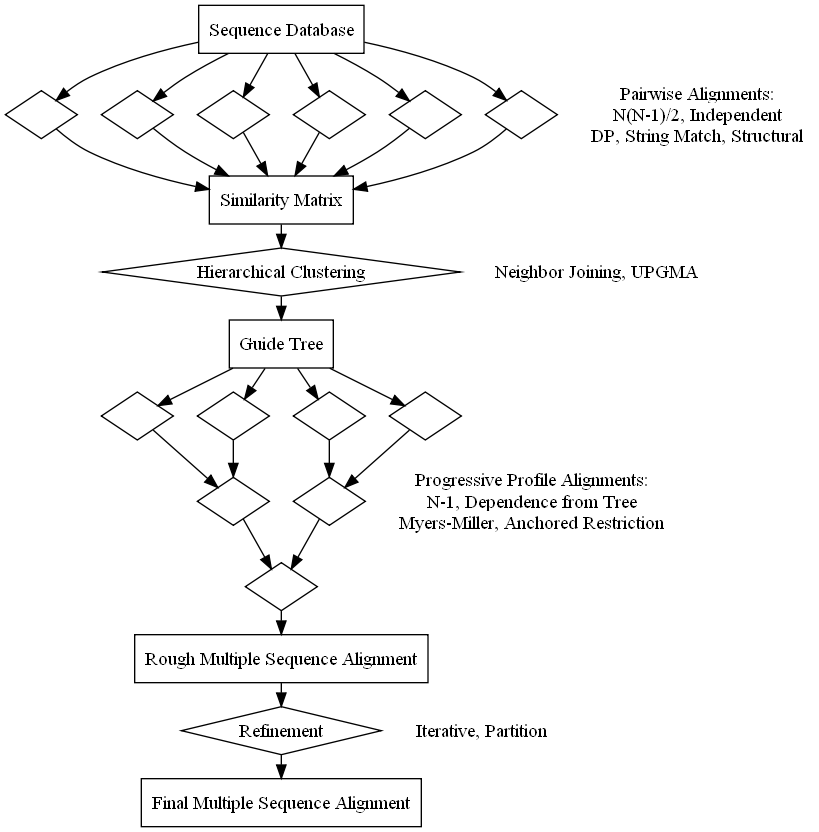
The first milestone consists of mostly design work, setting up package outlines, and writing interface hierarchies. The goal is to create an interface that allows both simple use with preset options and advanced customization. This will update the current alignment module to BioJava 3 and define the extension to multiple sequence alignments. The independence of the program components is key here. For example, pairwise alignments that rely on annotations of function or structure could create a similarity matrix while the rest of the routine defaults to run like clustalw. This flexible interface will allow the addition of more modern algorithms since most still follow the same four stage approach and even rely on many of the classic algorithms at some point during the alignment.
The second phase of this project implements a default alignment routine based on clustalw. Output from updated pairwise alignments will connect to the existing hierarchical clustering algorithms in the phylogeny module. Then, the resulting tree will guide a progressive alignment which outputs a multiple sequence alignment. This requires a data structure to hold profiles of multiple sequences and an algorithm for profile-profile alignment. Even the original release of clustalw uses several improvements to the naive algorithm which increase speed and accuracy.
For the remainder of the summer, enhancements will be added to each alignment stage. Many improvements upon clustalw have been published including clustalw2, muscle, kalign, dialign, mafft, fsa, probcons, and t-coffee, and psalign. Likely algorithms to integrate this summer are pairwise similarities from Needleman-Wunsch (clustalw), K-mers (clustalw/muscle), Wu-Manber (kalign), and structure (psc++/ce/fatcat), guide trees from Neighbor Joining and UPGMA, profile-profile alignment using a profile matrix (clustalw) or additive profile vectors (muscle) with anchored restriction (Myers-Miller/muscle/dialign), and refinement by both iterating (clustalw/muscle) and partitioning (muscle/kalign).
Timeline
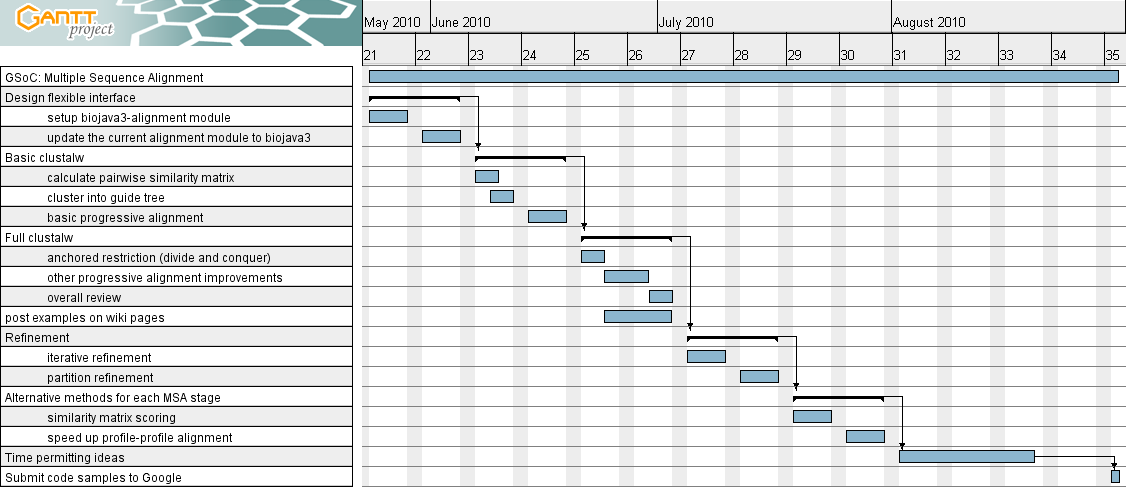
Design flexible interface
May 24 - June 4
- setup biojava3-alignment module
- package outline, interface hierarchies
- framework: standard four stage approach
- pairwise similarity calculation
- hierarchical clustering
- progressive alignment
- refinement
- submit for comments from development list
- code interface/class hierarchy
- mostly abstractions, stub methods with TODO comments, and
test cases
- example use case: pairwise structure alignments create similarity matrix while rest of the routine defaults to run like clustalw
- mostly abstractions, stub methods with TODO comments, and
test cases
- update the current alignment module to biojava3
- allow Compound list or string representation using BioJava 3 sequence package
- each alignment routine can choose either representation
- refine other aspects
- implement Iterable
wherever an iterator method exists - allows use of for-each loop through collection
- rework to better mesh with multiple sequence alignments
- implement Iterable
Basic clustalw
June 7 - 18
- calculate pairwise similarity matrix
- get scores from pairwise Needleman-Wunsch algorithm
- relatively slow, but already implemented in alignment module
- make multi-threaded version (default)
- all N*(N-1)/2 pairwise alignments are independent
- use ThreadPool utility, java.util.concurrent, Callable, Future…
- get scores from pairwise Needleman-Wunsch algorithm
- cluster into guide tree
- convert scores to normalized distances
- allow choice of Neighbor Joining or UPGMA clustering
- use implementations in phylo module
- NJ tree may need to be rooted
- basic progressive alignment
- naive profile-profile Needleman-Wunsch algorithm
- postfix tree traversal builds rough multiple sequence alignment
- make multi-threaded, list alignment calls from leaves to root
- produce profile scores by sum of pairs
Full clustalw
June 21 - July 2
- anchored restriction (divide and conquer)
- code Myers-Miller algorithm first
- generalize interface to later expand anchor sources (if time permits)
- other progressive alignment improvements
- use sequence weighting to combine similar and divergent
sequences
- adjusts for duplicate information that inflates sum of pairs scores
- choose substitution weight matrices
- strict for similar profiles and lenient for distant profiles
- PAM and BLOSUM series for amino acid substitution
- vary gap penalties for multiple reasons
- weight matrix: adjust opening penalty
- profile similarity: adjust opening penalty
- profile lengths: adjust opening penalty
- profile length differences: adjust extension penalty
- existing gaps: adjust opening and extension penalities
- specific residues: adjust opening penalty
- use sequence weighting to combine similar and divergent
sequences
- overall review
- check through design (especially public interface) for any improvements
- post examples on wiki pages
- show simple runs with default settings
- explain customization options and future modifications
Refinement
July 6 - July 16
- iterative refinement
- similarities from fractional identity of sequence pairs in rough MSA
- recluster, realign, repeat
- partition refinement
- split set of aligned sequences, realign
- single sequence removal, stochastic, and tree traversal
- split set of aligned sequences, realign
Alternative methods for each MSA stage
July 19 - 30
- similarity matrix scoring
- structure: psc++/ce/fatcat from structure module
- fast string matches: K-mers (clustalw/muscle), Wu-Manber
(kalign)
- UkkonenSuffixTree utility should help
- speed up profile-profile alignment
- additive profile vectors (muscle)
- more sources for anchored restriction (muscle/dialign)
Time permitting ideas
August 2 - 20
- polishing
- code: public interface, readability, extensibility
- JUnit tests: missing coverage, future possibilities
- documentation: JavaDoc comments and examples on wiki
- run benchmarks for protein (BAliBASE, PreFab) and RNA (BRAliBASE)
- update more code to BioJava3
- add scoring options
- CORE from T-Coffee
- accuracy/sensitivity/specificity/consistency/certainty from FSA
- check O(N^2) optimizations of NJ and UPGMA
- access clustr database for guide tree formation
- front end on PDB web site
- research and write roadmap for future directions
Submit code samples to Google
August 30
Progress Log
May 24 - May 28
- setup biojava3-alignment module
- read Effective Java by Joshua Bloch
- design work (incomplete)
- update to current alignment module
- extension to multiple sequence alignment
- ConcurrencyTools utility for submitting parallel tasks to a shared thread pool
June 1 - June 4
- design work
- posted to wiki
- integrated ideas from mentors
- started coding
June 7 - June 11
- implemented interface hierarchy
- added documentation comments
- outlined simple factory class Alignments
- implemented data structures
- gap penalty
- substitution matrix
- associated tests
June 14 - June 18
- implemented global pairwise sequence alignment
- aligned sequence
- profile
- sequence pair
- Needleman-Wunsch
- implemented parallelization
- concurrency tools: shared thread pool
- callable aligners and scorers
- factory methods which queue and run concurrent alignments and scorings
June 21 - June 25
- additional pairwise scoring options
- fractional identity
- fractional similarity
- guide tree formation
- convert scores into distance matrix
- call neighbor joining in forester library
- local pairwise sequence alignment
- factory method
- profile
- sequence pair
- Smith-Waterman
- profile-profile alignment
- aligned sequence
- profile
- profile pair
- abstract aligner
June 28 - July 2
- local pairwise sequence alignment
- added tests
- fixed bug in data structure
- similar/equivalent compounds
- implemented in amino acid compound set
- sparked discussion about compound sets
- provided example to user on mailing list
- alternative scorers
- added tests for identical compounds
- added tests for similar compounds
- profile-profile alignment
- refactored matrix aligners
- researched profile score functions
- started GuideTree wrapper class
July 5 - July 9
- profiles
- added column counts and weights
- added single sequence profiles
- profile-profile alignment
- refactored matrix aligners (minor)
- finished GuideTree wrapper class
- iterable from leaves to root
- finished naive implementation
- added caching of profile vectors
- added concurrent progressive MSA
- progressive MSA factory method
- allows easy multiple sequence alignments
July 12 - July 16
- rescore refinement
- FractionalIdentityInProfileScorer
- FractionalSimilarityInProfileScorer
- Profile from 2 AlignedSequence
- MSA options
- researched algorithms and scoring methods used in several common aligners
- listed stage options and ‘emulation’ defaults
- arranged Alignments static class in 3 access levels
- wrote simple example programs
- benchmarked time for MSA stages
July 19 - July 23
- posted simple example programs to biojava3 cookbook
- longer skype call than usual to prioritize additions yet this summer
- variable gap penalty
- linear space alignment
- read papers by Myers and Miller, Thompson, Guan and Uberbacher, and Morgenstern et al
- refinement
- benchmarking: time, memory, and quality
- downloaded the snoal and susd pfam families and the dengue and hiv genomes
- benchmarked time and memory (with 1.5GB limit)
- various improvements
- changed biojava3-genome dependency from jaligner to biojava3-alignment
- refactored AlignedSequence to have Sequence generic type
- fixed various compiler warnings in core module
- added distance and similarity methods to Scorer interface
- added new thread pool options as discussed in call
- disabled test in structure module to allow a successful build
- changed private methods in Alignments to default access
July 26 - July 30
- updated cookbook programs
- added them to test folder in repository
- added time and memory usage profiler
- made improvements to memory usage during multiple sequence alignment
- null out cached values after use (e.g. keep pairwise scores, but not alignments)
- various improvements
- added hasGap method to Profile; implemented in SimpleProfile
- removed MSAEmulation; added comments about similar routines at each stage
- worked around unchecked class cast warning; assert that compound sets match in sequences and substitution matrix
- provided simple access to common substitution matrices: SubstitutionMatrixHelper
- refactored aligner, scorer, and refiner enum types
August 2 - August 6
- survived strep throat
- decided to pursue
Guan-Uberbacher’s
linear space alignment
- simpler concept than Myers-Miller; only forward scoring passes
- allows multiple divisions in each pass; improves time requirement
- provides a hook for anchored alignments
August 9 - August 13
- refactored alignment routines
- granted access to all 3 score matrices built for affine gap penalties
- reduced memory requirement of single pass routines: score vectors and traceback matrix
- prepared for linear space alignment: traceback vectors (less memory), but multiple passes (more time)
- added alignment output formatting
- allows interlacing of sequences to show aligned columns
- combines a header, alignment information (indices, etc.), and sequence information (accession IDs, indices, etc.)
- outputs to CLUSTALW’s ALN, FASTA, and GCG’s MSF standard formats
Skype call notes
June 8th, June 15th, June 22nd, June 29th, July 21st, July 27th, Aug 10th, Aug 17th
References
- BioJava — <doi:10.1093/bioinformatics/btn397>
- Review — <doi:10.1093/bioinformatics/btp452>
- Clustalw2 — <doi:10.1093/bioinformatics/btm404>
- Clustalw — PMID: 7984417
- Clustal — PMID: 3243435
- Muscle — <doi:10.1186/1471-2105-5-113>
- Kalign — <doi:10.1186/1471-2105-6-298>
- Anchors — <doi:10.1186/1748-7188-1-6>
- Dialign — <doi:10.1186/1748-7188-3-6>
- T-Coffee — <doi:10.1006/jmbi.2000.4042>
- FSA — <doi:10.1371/journal.pcbi.1000392>
- ProbCons — <doi:10.1101/gr.2821705>
- BAliBASE — <doi:10.1002/prot.20527>
- Adaptive — <doi:10.1093/bioinformatics/bti828>
- Parallel — <doi:10.1186/1471-2105-5-128>
Comments
Please add comments here…